-
PDF
- Split View
-
Views
-
Cite
Cite
David Teze, Franck Daligault, Vincent Ferrières, Yves-Henri Sanejouand, Charles Tellier, Semi-rational approach for converting a GH36 α-glycosidase into an α-transglycosidase, Glycobiology, Volume 25, Issue 4, April 2015, Pages 420–427, https://doi.org/10.1093/glycob/cwu124
Close - Share Icon Share
Abstract
A large number of retaining glycosidases catalyze both hydrolysis and transglycosylation reactions. In order to use them as catalysts for oligosaccharide synthesis, the balance between these two competing reactions has to be shifted toward transglycosylation. We previously designed a semi-rational approach to convert the Thermus thermophilus β-glycosidases into transglycosidases by mutating highly conserved residues located around the −1 subsite. In an attempt to verify that this strategy could be a generic approach to turn glycosidases into transglycosidases, Geobacillus stearothermophilus α-galactosidase (AgaB) was selected in order to obtain α-transgalactosidases. This is of particular interest as, to date, there are no efficient α-galactosynthases, despite the considerable importance of α-galactooligosaccharides. Thus, by site-directed mutagenesis on 14 AgaB residues, 26 single mutants and 22 double mutants were created and screened, of which 11 single mutants and 6 double mutants exhibited improved synthetic activity, producing 4-nitrophenyl α-d-galactopyranosyl-(1,6)-α-d-galactopyranoside in 26–57% yields against only 22% when native AgaB was used. It is interesting to note that the best variant was obtained by mutating a second-shell residue, with no direct interaction with the substrate or a catalytic amino acid. As this approach has proved to be efficient with both α- and β-glycosidases, it is a promising route to convert retaining glycosidases into transglycosidases.
Introduction
Carbohydrates play major roles in most biological processes such as inflammation reactions, cell death, cellular interactions, stabilization of proteins, adhesion of pathogenic microorganisms and binding of toxins (Hart and Copeland 2010; Ghazarian et al., 2011; Lichtenstein and Rabinovich 2013). In particular, most naturally occurring human antibodies recognize a carbohydrate, the α-gal epitope (Macher and Galili 2008) and α-galactosyl residues are present in glycan antigens targeted by carbohydrate vaccines against various pathogens such as Neisseria meningitidis, Shigella dysenteriae and Leishmania spp. (Astronomo and Burton 2010). Furthermore, α-1,6-linked galactooligosaccharides have been recognized for their prebiotic properties (Nakai et al. 2010; Cervera-Tison et al. 2012; Hachem et al. 2012; Andersen et al. 2013; Wang et al. 2014). However, their use is hindered by their poor availability: Unlike for proteins and nucleic acids, there is no general synthetic route for oligosaccharides and, despite the considerable development of efficient methods in this field (Wang et al. 2007; Zhu and Schmidt 2009), the assembly of oligosaccharides remains a substantial challenge. Consequently, alternative synthetic strategies have been explored, particularly those based on enzymatic processes.
The enzymatic synthesis of glycosidic bonds can be performed by glycosyltransferases or transglycosidases. The former can catalyze the formation of glycosidic bonds with both high yield and selectivity, but their use is limited in the large-scale production of oligosaccharides by the difficulty in accessing activated sugar donors (sugar nucleotides for Leloir-type transferases or phosphate sugars for non-Leloir types) (Lairson et al. 2008), even though a recent approach seems promising (Zhang et al. 2006; Gantt et al. 2013). As an alternative, engineered retaining glycosidases have provided an efficient approach for the synthesis of oligosaccharides. A first methodology was based on the substitution of the catalytic nucleophile by a neutral amino acid and the use of activated donors with an anomeric configuration opposite to that of the original substrate (such as α-glycosyl fluorides), which yielded new enzymes called glycosynthases (Wang et al. 1994; Mackenzie et al. 1998; Malet and Planas 1998; Cobucci-Ponzano et al. 2012). However, although these glycosynthases have been very efficient for the synthesis of β-glycosidic bonds, the results are disappointing when they are applied to α-galactosidases, despite recent advances (Cobucci-Ponzano et al. 2010). A second methodology based on the Hehre-resynthesis hydrolysis properties of inverting glycosidases (Hehre et al. 1979) has been proposed. Mutating residues involved in the hydrolysis reaction but not in the Hehre-resynthesis reaction yield variants that efficiently catalyze the synthesis β-linkages (Honda and Kitaoka 2006; Honda et al. 2008). This methodology was also extended to the synthesis of α-linkages (Wada et al. 2008), but with less efficiency.
As an alternative, natural α-transglucosidases, such as amylosucrase and dextransucrase, have been identified in the Carbohydrate-Active Enzymes Database (CAZy) (Cantarel et al. 2009) families GH13 and GH70 that can efficiently catalyze the formation of α-glucosidic bonds using sucrose as a natural activated donor (Pizzut-Serin et al. 2005; Plou et al. 2007; Champion et al. 2009; Irague et al. 2011). These examples of natural transglycosidases suggested that directed evolution of glycosidases into transglycosidases could be possible and this strategy was validated in our laboratory on both α- and β-glycosidases (Feng et al. 2005; Osanjo et al. 2007; Koné et al. 2009). Although molecular modeling and structural analysis were tentatively applied to explain why these evolved mutants present improved transglycosidase activities (Feng et al. 2005; Osanjo et al. 2007; Teze and Hendrickx 2013; Teze et al. 2014), the molecular events leading to an improved transglycosylation over hydrolysis ratio have not yet been determined precisely, even for natural transglycosidases such as cyclodextrin glycosyltransferases (Leemhuis et al. 2002; Kelly et al. 2008). However, the data led us to propose the working hypothesis that mutating conserved residues within the donor subsite of Thermus thermophilus β-glycosidase would enable the creation of transglycosidases and indeed four new β-transglycosidases were obtained (Teze et al. 2014).
Therefore, the aim of this study was to investigate whether this approach could also enable the rapid creation of efficient α-transgalactosidases. AgaB was chosen as a model system as this enzyme, which belongs to the CAZy family 36 (Lombard et al. 2014), subgroup I (Fredslund et al. 2011; Hachem et al. 2012), has shown interesting transgalactosylation properties. It was shown to catalyze the synthesis of 4-nitrophenyl α-d-galactopyranosyl-(1,6)-α-d-galactopyranoside (GalGal-pNP) with yields up to 53% when high substrate concentrations were used (100 mM) (Spangenberg et al. 2000). Higher yields of this disaccharide (74% with 40 mM of substrate) have been obtained with the α-galactosidase from Aspergillus nidulans, which belongs to the same subgroup and shares 39% identity with AgaB (Nakai et al. 2010). We also previously showed that the regioselectivity of the enzyme could be modulated by mutagenesis (Dion and Nisole 2001; Dion and Osanjo 2001). In addition, its crystallographic structure has recently been published (Merceron et al. 2012). Recently, the first reported α-galactosynthase was created from a GH36 enzyme, the Thermotoga maritima α-galactosidase (TmαGal, subgroup III) (Cobucci-Ponzano et al. 2010) and moderate improvement of the transglycosylation catalyzed by the same enzyme has been reported (Bobrov et al. 2013). Interestingly, the GH36 family also contains pure transgalactosidases, such as stachyose and verbascose synthases, which are important for carbon storage in seeds (Peterbauer and Mach 2002; Peterbauer and Mucha 2002; Peterbauer et al. 2003). However, these plant enzymes, which belong to the GH36 subgroup II (Fredslund et al. 2011), are difficult to produce and their structure is not known. Their sequences present <15% identity with AgaB, preventing reliable sequence alignments, not to mention homology modeling. Nevertheless, their existence suggests that glycoside hydrolases from this family could be evolved to pure transglycosidases, with little or no hydrolysis activity, while keeping their original double-displacement mechanism.
The present work describes a semi-rational approach to convert the Geobacillus stearothermophilus α-galactosidase (AgaB) into transglycosidases by mutating highly conserved residues located around the −1 donor subsite. An initial Ala scanning allowed identifying critical positions that affect the hydrolysis/tranglycosylation ratio, and the most effective positions were then mutated into a more conservative amino acid. We found that 50% of the mutated positions led to an improvement of the transglycosidase activity, suggesting that targeting the −1 site of glycosidases could significantly decrease the screening effort to evolve glycosidases into tranglycosidases.
Results
Amino acid conservation within the GH36 family
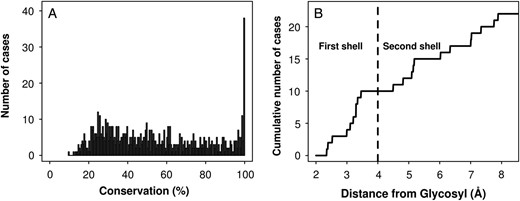
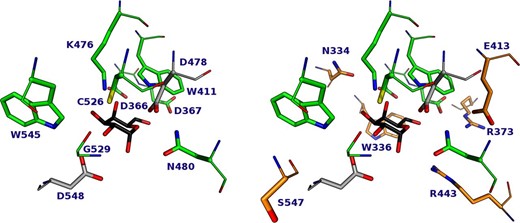
To study the effect of mutations on conserved residues within the donor subsite of the enzyme, the conservation of all AgaB amino acids was investigated. Thus, using the AgaB sequence as the query, 205 sequences from the GH36 subgroup I were obtained that shared between 25 and 95% of sequence identity between all pairs of sequences and between 25 and 66% with the query sequence. The average conservation was 54.4%, with 5.2% of amino acids (38 AAs) at least 99.5% conserved (Figure 1A). Given the AgaB structure [protein data bank (PDB) code 4FNQ] and the very homologous AgaA structure (97% identity) in complex with stachyose (PDB code 4FNU) (Merceron et al. 2012), an α-Gal residue was added to the AgaB structure by superimposition (root mean square deviation = 0.5 Å). The minimal distance between one of the nonhydrogen atoms of the aforementioned conserved residues and one of the glycosyl nonhydrogen atoms was measured (Figure 1B). It is particularly interesting to note that 10 residues have a minimal distance <3.5 Å from the glycosyl, and that no other residues have a minimal distance <4.5 Å. These 10 residues constitute the first shell, directly involved in catalytic activity (D478 and D548) or in substrate binding (D366, D367, W411, K476, N480, C526, G529 and W545; Figure 2A). Six of these highly conserved residues are located from 4.5 to 6.1 Å away from the glycosyl, interacting with first-shell residues and thus constituting the conserved residues of the second interaction shell (N334, W336, R373, E413, R443 and S547; Figure 2B). These 16 residues located at <6.1 Å from the glycosyl are likely to be involved in the enzyme function and their impact on the transglycosylation is worth exploring. Then, the next conserved residue is the G368 at 7 Å, which might be involved in protein folding.
Amino acid conservation within the GH36 family. (A) Histogram of the population of amino acids as a function of their percentage of conservation. Each population corresponds to an interval of 0.5% of conservation and the statistics are based on 205 sequences. (B) Highly conserved amino acid population as a function of the distance from the glycosyl, based on the AgaB structure (PDB code 4FNQ).
Localization of highly conserved amino acids within the AgaB active site (PDB code 4FNQ). Left: First-shell residues. Right: First and second-shell residues. Only Second-shell residues are labeled. This figure was created with PyMol.
Ala-scanning
Among the 16 conserved AAs close to the donor subsite, two are the catalytic residues (D478 and D548) and the remaining 14 were individually mutated into alanine to assess if these residues have an influence on the transglycosylation yields. The resulting mutants were produced and their kinetic parameters kcat/KM as well as their ability to catalyze GalGal-pNP synthesis are summarized in Table I.
Kinetics and yields of GalGal-pNP synthesis of the AgaB variants obtained by Ala-scanning
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WT | 200 ± 4 | 22 | 1.0 |
| G529A | 3.06 ± 0.03 | 21 | 5.5 |
| W545A | (6 ± 0.3) × 10−2 | 34 | 790 |
| C526A | 0.33 ± 0.02 | 19 | 34 |
| S547A | 0.70 ± 0.05 | 31 | 29 |
| N334A | 0.57 ± 0.02 | 38 | 170 |
| R373A | 0.67 ± 0.04 | 2 | 11 |
| W411Ab | (1.3 ± 0.1) × 10−2 | NDc | NDc |
| E413Ab | (1.2 ± 0.1) × 10−2 | NDc | NDc |
| K476A | (3.1 ± 0.1) × 10−3 | NDc | NDc |
| W336A; R443A | NDd | NDc | NDc |
| N480A; D366A; D367A | NDAe | NDc | NDc |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WT | 200 ± 4 | 22 | 1.0 |
| G529A | 3.06 ± 0.03 | 21 | 5.5 |
| W545A | (6 ± 0.3) × 10−2 | 34 | 790 |
| C526A | 0.33 ± 0.02 | 19 | 34 |
| S547A | 0.70 ± 0.05 | 31 | 29 |
| N334A | 0.57 ± 0.02 | 38 | 170 |
| R373A | 0.67 ± 0.04 | 2 | 11 |
| W411Ab | (1.3 ± 0.1) × 10−2 | NDc | NDc |
| E413Ab | (1.2 ± 0.1) × 10−2 | NDc | NDc |
| K476A | (3.1 ± 0.1) × 10−3 | NDc | NDc |
| W336A; R443A | NDd | NDc | NDc |
| N480A; D366A; D367A | NDAe | NDc | NDc |
aEnzyme concentration necessary to reach the maximum yield of GalGal-pNP formation within 4 h.
bW411A (kcat = (1.9 ± 0.02) × 10−2 s−1; KM = 1.5 ± 0.07 mM) and E413A (kcat = (3.0 ± 0.1) × 10−2 s−1; KM = 2.4 ± 0.2 mM) display a classic Michaelis–Menten behavior.
cNot determined. Activities of these enzymes were too low to measure GalGal-pNP formation.
dW336A and R443A presented a detectable activity, but too low to be measured.
eNo detectable activity.
Kinetics and yields of GalGal-pNP synthesis of the AgaB variants obtained by Ala-scanning
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WT | 200 ± 4 | 22 | 1.0 |
| G529A | 3.06 ± 0.03 | 21 | 5.5 |
| W545A | (6 ± 0.3) × 10−2 | 34 | 790 |
| C526A | 0.33 ± 0.02 | 19 | 34 |
| S547A | 0.70 ± 0.05 | 31 | 29 |
| N334A | 0.57 ± 0.02 | 38 | 170 |
| R373A | 0.67 ± 0.04 | 2 | 11 |
| W411Ab | (1.3 ± 0.1) × 10−2 | NDc | NDc |
| E413Ab | (1.2 ± 0.1) × 10−2 | NDc | NDc |
| K476A | (3.1 ± 0.1) × 10−3 | NDc | NDc |
| W336A; R443A | NDd | NDc | NDc |
| N480A; D366A; D367A | NDAe | NDc | NDc |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WT | 200 ± 4 | 22 | 1.0 |
| G529A | 3.06 ± 0.03 | 21 | 5.5 |
| W545A | (6 ± 0.3) × 10−2 | 34 | 790 |
| C526A | 0.33 ± 0.02 | 19 | 34 |
| S547A | 0.70 ± 0.05 | 31 | 29 |
| N334A | 0.57 ± 0.02 | 38 | 170 |
| R373A | 0.67 ± 0.04 | 2 | 11 |
| W411Ab | (1.3 ± 0.1) × 10−2 | NDc | NDc |
| E413Ab | (1.2 ± 0.1) × 10−2 | NDc | NDc |
| K476A | (3.1 ± 0.1) × 10−3 | NDc | NDc |
| W336A; R443A | NDd | NDc | NDc |
| N480A; D366A; D367A | NDAe | NDc | NDc |
aEnzyme concentration necessary to reach the maximum yield of GalGal-pNP formation within 4 h.
bW411A (kcat = (1.9 ± 0.02) × 10−2 s−1; KM = 1.5 ± 0.07 mM) and E413A (kcat = (3.0 ± 0.1) × 10−2 s−1; KM = 2.4 ± 0.2 mM) display a classic Michaelis–Menten behavior.
cNot determined. Activities of these enzymes were too low to measure GalGal-pNP formation.
dW336A and R443A presented a detectable activity, but too low to be measured.
eNo detectable activity.
Most of the mutants, except for the E413A and W411A variants, display a non-Michaelian behavior as they do not present any saturation at high substrate concentration, thus catalytic constants, KM and kcat, could not be individually determined and only the apparent second-order rate constant kcat/KM (“catalytic efficiency”) is presented (Table I). The wild-type (WT) enzyme and the G529A showed a slight substrate inhibition. Examples of these nonclassical behaviors are presented in Supplementary Data.
Of the 14 mutants produced, only 6 displayed sufficient activity to measure their yield of GalGal-pNP synthesis when used with α-Gal-pNP 10 mM as a substrate (Table I). This Ala-scanning was successful for rapidly obtaining transglycosidases, with three variants presenting transglycosylation yields in the range 31–38% against 22% with the WT. However, the kinetic parameter kcat/KM of these six mutants dropped dramatically by two to three orders of magnitude. It should be noted that in three (G529A, C526A and S547A) of these six variants, Ala mutation resulted in low steric change and that two of these three presented the highest activities among all variants. While Ala-scanning was performed to analyze the effect of each conserved residue substitution in a similar fashion, it appeared that minimizing the steric difference between the variant and the native enzyme was needed in order to avoid drastic loss of activity. Thus, a second round of mutagenesis featuring “conservative mutations,” that is, mutations closest in structure to the WT enzyme, was performed.
Conservative mutations
Of the 14 chosen positions, 10 were selected for a second round of mutagenesis in which the novel amino acid replaced the original one in a more isosteric way. G529, C526 and S547 were excluded as Ala is already a good replacement choice and mutating R373 was excluded as it seemed to engender a strong deleterious effect on the transglycosylation yield, with a drastic drop from 22 to 2% (Table I). As the N334A mutant was the best variant selected by Ala-scanning, more substitutions on this specific position were added. The kinetic parameters kcat/KM as well as GalGal-pNP synthesis yields of the resulting mutants are summarized in Table II.
Kinetics and yields of GalGal-pNP synthesis of AgaB variants presenting “conservative mutations”
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334D | 6.6 ± 0.2 | 44 | 3.0 |
| N334S | 2.4 ± 0.02 | 39 | 11.5 |
| N334T | 5.4 ± 0.03 | 45 | 10.5 |
| W336F | 0.38 ± 0.005 | 27 | 205 |
| D367Nb | 0.23 ± 0.005 | 26 | 55 |
| W411Fb | 0.75 ± 0.02 | 27 | 23 |
| K476Q | (6.7 ± 0.4) × 10−2 | 36 | 115 |
| N480D | (7.3 ± 0.3) × 10−2 | 14 | 140 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| R443Q; D366N; E413Q | NDAc | NDA | NDA |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334D | 6.6 ± 0.2 | 44 | 3.0 |
| N334S | 2.4 ± 0.02 | 39 | 11.5 |
| N334T | 5.4 ± 0.03 | 45 | 10.5 |
| W336F | 0.38 ± 0.005 | 27 | 205 |
| D367Nb | 0.23 ± 0.005 | 26 | 55 |
| W411Fb | 0.75 ± 0.02 | 27 | 23 |
| K476Q | (6.7 ± 0.4) × 10−2 | 36 | 115 |
| N480D | (7.3 ± 0.3) × 10−2 | 14 | 140 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| R443Q; D366N; E413Q | NDAc | NDA | NDA |
aEnzyme concentration necessary to reach the maximum amount of GalGal-pNP within 4 h.
bThese AgaB variants present a slight substrate inhibition.
cNo detectable activity.
Kinetics and yields of GalGal-pNP synthesis of AgaB variants presenting “conservative mutations”
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334D | 6.6 ± 0.2 | 44 | 3.0 |
| N334S | 2.4 ± 0.02 | 39 | 11.5 |
| N334T | 5.4 ± 0.03 | 45 | 10.5 |
| W336F | 0.38 ± 0.005 | 27 | 205 |
| D367Nb | 0.23 ± 0.005 | 26 | 55 |
| W411Fb | 0.75 ± 0.02 | 27 | 23 |
| K476Q | (6.7 ± 0.4) × 10−2 | 36 | 115 |
| N480D | (7.3 ± 0.3) × 10−2 | 14 | 140 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| R443Q; D366N; E413Q | NDAc | NDA | NDA |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334D | 6.6 ± 0.2 | 44 | 3.0 |
| N334S | 2.4 ± 0.02 | 39 | 11.5 |
| N334T | 5.4 ± 0.03 | 45 | 10.5 |
| W336F | 0.38 ± 0.005 | 27 | 205 |
| D367Nb | 0.23 ± 0.005 | 26 | 55 |
| W411Fb | 0.75 ± 0.02 | 27 | 23 |
| K476Q | (6.7 ± 0.4) × 10−2 | 36 | 115 |
| N480D | (7.3 ± 0.3) × 10−2 | 14 | 140 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| R443Q; D366N; E413Q | NDAc | NDA | NDA |
aEnzyme concentration necessary to reach the maximum amount of GalGal-pNP within 4 h.
bThese AgaB variants present a slight substrate inhibition.
cNo detectable activity.
In the set of variants obtained by “conservative mutations,” only three mutants were inactive: R443Q, D366N, E413Q and a 10- to 75-fold increase in kcat/KM was observed on four positions with regard to Ala mutants as well as a 15- to 100-fold decrease in the amount of enzyme needed to reach reaction completion. Consequently, eight of nine variants were active enough to assay GalGal-pNP synthesis and showed improved yields compared with the native enzyme (from 26 to 45% against 22% with the WT). Interestingly, none of the newly created variants displayed yields lower than the corresponding Ala variant, which suggest that starting directly with a round of conservative mutations is most probably the best approach to create new transglycosidases.
As the resulting mutants after this second round of mutagenesis kept high levels of activity, combinations of the most successful mutations were performed.
Mutant recombination
After two rounds of mutagenesis, the variants obtained by mutating conserved residues of 7 of 14 positions showed an increase in transglycosylation yields, therefore 21 recombination mutants, resulting from the combination of mutations S547A, N334T, W336F, D267N, W411F, K476Q and W545F, were created. The W545F-N334S double mutant was added to this set. Kinetic parameters kcat/KM as well as GalGal-pNP synthesis yield of the resulting mutants are summarized in Table III.
Kinetic parameters and yields of GalGal-pNP synthesis of the double mutants obtained by recombination
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334Tb | 5.4 ± 0.03 | 45 | 10.5 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| N334T-W545F | 0.21 ± 0.02 | 57 | 360 |
| W545F-N334S | 0.12 ± 0.004 | 40 | 425 |
| W545F-W411F | (1.1 ± 0.1) × 10−2 | 44 | 1.7 × 103 |
| N334T-W411F | (3.6 ± 0.3) × 10−2 | 44 | 280 |
| W545F-W336F | (3.8 ± 0.1) × 10−2 | 26 | 1.0 × 103 |
| N334T-K476Q | (1.9 ± 0.2) × 10−2 | 38 | 1.4 × 103 |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334Tb | 5.4 ± 0.03 | 45 | 10.5 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| N334T-W545F | 0.21 ± 0.02 | 57 | 360 |
| W545F-N334S | 0.12 ± 0.004 | 40 | 425 |
| W545F-W411F | (1.1 ± 0.1) × 10−2 | 44 | 1.7 × 103 |
| N334T-W411F | (3.6 ± 0.3) × 10−2 | 44 | 280 |
| W545F-W336F | (3.8 ± 0.1) × 10−2 | 26 | 1.0 × 103 |
| N334T-K476Q | (1.9 ± 0.2) × 10−2 | 38 | 1.4 × 103 |
aEnzyme concentration necessary to reach the maximum amount of GalGal-pNP within 4 h.
bWT, N334T and W545F were included as a reminder of the effect of single mutations.
Kinetic parameters and yields of GalGal-pNP synthesis of the double mutants obtained by recombination
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334Tb | 5.4 ± 0.03 | 45 | 10.5 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| N334T-W545F | 0.21 ± 0.02 | 57 | 360 |
| W545F-N334S | 0.12 ± 0.004 | 40 | 425 |
| W545F-W411F | (1.1 ± 0.1) × 10−2 | 44 | 1.7 × 103 |
| N334T-W411F | (3.6 ± 0.3) × 10−2 | 44 | 280 |
| W545F-W336F | (3.8 ± 0.1) × 10−2 | 26 | 1.0 × 103 |
| N334T-K476Q | (1.9 ± 0.2) × 10−2 | 38 | 1.4 × 103 |
| Enzyme . | kcat/KM (s−1 mM−1) . | GalGal-pNP (%) . | Enzyme concentration (µg mL−1)a . |
|---|---|---|---|
| WTb | 200 ± 4 | 22 | 1.0 |
| N334Tb | 5.4 ± 0.03 | 45 | 10.5 |
| W545Fb | 2.7 ± 0.02 | 35 | 7.5 |
| N334T-W545F | 0.21 ± 0.02 | 57 | 360 |
| W545F-N334S | 0.12 ± 0.004 | 40 | 425 |
| W545F-W411F | (1.1 ± 0.1) × 10−2 | 44 | 1.7 × 103 |
| N334T-W411F | (3.6 ± 0.3) × 10−2 | 44 | 280 |
| W545F-W336F | (3.8 ± 0.1) × 10−2 | 26 | 1.0 × 103 |
| N334T-K476Q | (1.9 ± 0.2) × 10−2 | 38 | 1.4 × 103 |
aEnzyme concentration necessary to reach the maximum amount of GalGal-pNP within 4 h.
bWT, N334T and W545F were included as a reminder of the effect of single mutations.
Among the 22 newly created double mutants, only 6 were active enough to allow quantification of GalGal-pNP synthesis (Table III), and these 6 mutants always contained W545F and/or N334T mutations, which are the least deleterious to the kcat/KM parameter among those used for recombination (Tables I and II). It can be concluded from these 15 inactive or low active mutants that there is a strong additivity of the decrease in catalytic efficiency, while the beneficial effect on transglycosylation shows a less additive effect: Only the double mutant N334T-W545F displayed a higher yield than the single mutant N334T or W545F, with a 57% yield (against 45% for N334T, 35% for W545F and 22% for the WT).
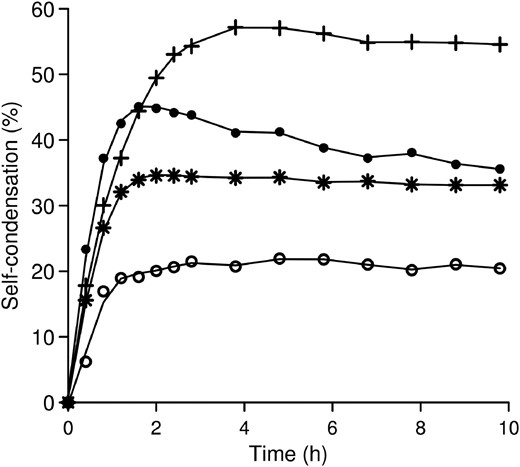
For the best variants (N334T, W545F and N334T-W545F), the kinetics of GalGal-pNP synthesis were compared with that of the WT (Figure 3). One of the drawbacks of the use of glycosidases for oligosaccharide synthesis is often the secondary hydrolysis of the formed products. However, thanks to the use of an activated donor for which AgaB presents a much higher specificity than for natural substrates (the kcat/KM toward α-Gal-pNP is 500 times greater than toward raffinose (Merceron et al. 2012)), small amounts of enzyme were used and the transglycosylation product was barely hydrolyzed and maximum yields of product formation remained for a long time (Figure 3). Only the N334T variant exhibited a slight secondary hydrolysis compared with the other mutants and the native enzyme.
Time course of GalGal-pNP synthesis in the presence of α-Gal-pNP 10 mM as substrate. Percentages are calculated with reference to initial substrate concentration using the following AgaB variants: WT (empty circles); W545F (stars); N334T (filled circles) and N334T-W545F (plus symbols).
After these three rounds of mutagenesis, AgaB variants with a strong improvement in their synthetic activity toward GalGal-pNP synthesis were obtained. However, as 4-nitrophenyl groups are hard to remove without breaking the glycosidic bond, this disaccharide is of little interest. Thus, a study of transglycosylation toward an acceptor bearing a thiophenyl group instead was undertaken, as this protecting group has proved successful in chemo-enzymatic synthesis (Marton et al. 2008; D'Almeida et al. 2009).
Effect of changes in donors and acceptors on synthetic yields of transglycosylation with AgaB variants
The closest analog of α-Gal-pNP containing a thiophenyl group is phenyl 1-thio-α-d-galactopyranoside. However, given the low availability of this compound, the more affordable phenyl 1-thio-β-d-galactopyranoside (Gal-S-Ph) was used. Moreover, this compound made it possible to assess whether the N334T variant could use acceptors with a β-configuration as well as with an α-configuration. The N334T variant was chosen over the N334T-W545F double mutant for the sake of enzyme consumption, as it gives the best transglycosylation yield after the N334T-W545F variant, but needs 35-fold less enzyme to reach reaction completion (Table IV). Transglycosylation yields of phenyl α-d-galactopyranosyl-(1,6)-1-thio-β-d-galactopyranoside (GalGal-S-Ph) synthesis are given in Table IV. Both native AgaB and the N334T variant were able to catalyze the synthesis of GalGal-S-Ph, with a net increase from 14 to 33% with the latter, which demonstrates that newly created variants keep the promiscuity of the native enzyme toward acceptors. A comparative study with the α-galactosynthase approach (Cobucci-Ponzano et al. 2010) has been also performed: The D478G AgaB variant (equivalent to the D327G TmαGal glycosynthase variant) was produced and tested with β-Gal-N3 as donor. It showed an interesting increase in synthetic yields compared with the native enzyme (from 14 to 23%, Table IV). Nevertheless, the α-transgalactosidase N334T variant displayed higher yields (33 against 23%), while consuming 42-fold less enzyme. When using Gal-S-Ph instead of α-Gal-pNP as an acceptor, a drop of one-third in synthetic yields with both N334T and the native enzyme was observed. This decrease is mainly due to competition with the GalGal-pNP synthesis. This competition could be turned toward synthesis by adding α-Gal-pNP gradually, which resulted in an increase from 33 to 54% of GalGal-S-Ph synthesis yield when the N334T variant was used. In order to go further in avoiding competition with the GalGal-pNP synthesis, α-Gal-F was used as donor instead of α-Gal-pNP (Andre et al. 2001). This resulted in a drastic increase in transglycosylation yields from 14 to 37% with the native enzyme and from 33 to 62% with the N334T variant (Table IV).
Yields of GalGal-S-Ph synthesis using AgaB variants with Gal-S-Ph (10 mM) as acceptor and various donors (each at 10 mM)
| Enzyme . | Donor . | GalGal-pNP (%)a . | Transglycosylation yield (%) . | Enzyme concentration (µg mL−1)b . |
|---|---|---|---|---|
| WT | α-Gal-pNP | 9 | 14 | 1.5 |
| WT | α-Gal-F | – | 37 | 0.3 |
| N334T | α-Gal-pNP | 19 | 33 | 18 |
| N334T | α-Gal-pNPc | 9 | 54 | 27 |
| N334T | α-Gal-F | – | 62 | 5.0 |
| D478G | β-Gal-N3 | 0 | 23 | 755 |
| Enzyme . | Donor . | GalGal-pNP (%)a . | Transglycosylation yield (%) . | Enzyme concentration (µg mL−1)b . |
|---|---|---|---|---|
| WT | α-Gal-pNP | 9 | 14 | 1.5 |
| WT | α-Gal-F | – | 37 | 0.3 |
| N334T | α-Gal-pNP | 19 | 33 | 18 |
| N334T | α-Gal-pNPc | 9 | 54 | 27 |
| N334T | α-Gal-F | – | 62 | 5.0 |
| D478G | β-Gal-N3 | 0 | 23 | 755 |
aYields of GalGal-pNP synthesis, measured at the time of the maximum yield of the transglycosylation product.
bEnzyme concentration necessary to reach the maximum transglycosylation yield within 4 h.
cInstead of having 10 mM of donor at the beginning of the reaction, the same amount of donor was reached by 5 additions during the time course of the reaction.
Yields of GalGal-S-Ph synthesis using AgaB variants with Gal-S-Ph (10 mM) as acceptor and various donors (each at 10 mM)
| Enzyme . | Donor . | GalGal-pNP (%)a . | Transglycosylation yield (%) . | Enzyme concentration (µg mL−1)b . |
|---|---|---|---|---|
| WT | α-Gal-pNP | 9 | 14 | 1.5 |
| WT | α-Gal-F | – | 37 | 0.3 |
| N334T | α-Gal-pNP | 19 | 33 | 18 |
| N334T | α-Gal-pNPc | 9 | 54 | 27 |
| N334T | α-Gal-F | – | 62 | 5.0 |
| D478G | β-Gal-N3 | 0 | 23 | 755 |
| Enzyme . | Donor . | GalGal-pNP (%)a . | Transglycosylation yield (%) . | Enzyme concentration (µg mL−1)b . |
|---|---|---|---|---|
| WT | α-Gal-pNP | 9 | 14 | 1.5 |
| WT | α-Gal-F | – | 37 | 0.3 |
| N334T | α-Gal-pNP | 19 | 33 | 18 |
| N334T | α-Gal-pNPc | 9 | 54 | 27 |
| N334T | α-Gal-F | – | 62 | 5.0 |
| D478G | β-Gal-N3 | 0 | 23 | 755 |
aYields of GalGal-pNP synthesis, measured at the time of the maximum yield of the transglycosylation product.
bEnzyme concentration necessary to reach the maximum transglycosylation yield within 4 h.
cInstead of having 10 mM of donor at the beginning of the reaction, the same amount of donor was reached by 5 additions during the time course of the reaction.
Discussion
With the best N334T AgaB variant, one-step glycosidic bond formation with high yields (from 45 to 62%) with unprotected sugars and moderate enzyme consumption was achieved, which makes this variant suitable for bioconversion uses. It also competes favorably with the glycosynthase D478G variant as it provides higher transglycosylation yields (62 against 23%), while using 151-fold less enzyme (Table IV). Thus, this approach, which targets highly conserved amino acids around the active site of AgaB, is very efficient for obtaining transglycosidase mutants rapidly and avoiding the tedious screening procedures imposed by random evolution approaches (Koné et al. 2009). Furthermore, the initial approach (Teze et al. 2014) has been improved, as we demonstrate in this work that one round of conservative mutations allowed a fast access to new transglycosidases while avoiding drastic drops in activity. It is particularly interesting to note that the best variant, N334T, was obtained by mutating a second-shell residue, showing that very slight modifications may provoke strong effects on the balance of transglycosylation over hydrolysis. The addition of a second mutation induced a strong deleterious effect on the activity of all the 22 double mutants produced (Table III). On the other hand, the effect on transglycosylation yields seems less predictable: Among the six double mutants that exhibited an activity high enough to study the transglycosylation yields, two displayed an interesting increase compared with the single mutants [N334T-W545F (+12%) and W411F-W545F (+9%)], two showed slight yield modifications [N334S-W545F (+1%) and N334T-W411F (−1%)] and two presented a drastic drop in yields [N334T-K476Q (−7%) and W336F-W545F (−9%)] (Table III).
Mutants were screened for the best transglycosidase activity by following the GalGal-pNP synthesis. As this product is of limited interest as an intermediate in oligosaccharide synthesis, we also demonstrated that by switching to other acceptors, mutants kept their improved transglycosylation properties since mutations did not affect the +1 and +2 acceptor sites. Moreover, by using alternative donors such as α-Gal-F, the transglycosylation yields improved due to the absence of the competing GalGal-pNP synthesis. As the α-Gal-F is a more activated donor than the α-Gal-pNP, the enzyme consumption was decreased by 3.6- to 5-fold, which also contributes to the increase in transglycosylation yields due to the reduction of secondary hydrolysis.
The spatial arrangement of the mutated residues can be seen in the published structure of AgaB (Merceron et al. 2012) (Figure 2). The N480 residue is located in the neighborhood of the acceptor subsite, which may explain the drop in transglycosylation yields obtained with the variant N480D as some interactions with an acceptor moiety could be missing. However, all the most beneficial mutations for transglycosidase activity are located farther from the catalytic residues, at the bottom of the −1 site cavity [mutations on residues S547, N334, W545 and K476 (Figure 2)]. Thus, structural information does not give any clues for understanding the effect of mutation on the transglycosylation/hydrolysis ratio. To date, a possible explanation is that highly conserved amino acids in the strict vicinity of the glycosyl subsite are involved in stabilizing the transition states of the catalyzed reactions, that is, glycosylation, hydrolysis and transglycosylation. Thus, mutating these residues might decrease the transition state stabilization of these three reactions (resulting in a drop in kinetic parameters), but their contributions to the stabilization of each of these three transition states are most probably not identical. Thus, the finely tuned equilibrium between these three reactions would be disturbed and it is likely that variants presenting a balance slightly shifted toward transglycosylation would be obtained (as well as those with a balance shifted toward hydrolysis). Indeed, on 14 mutated positions, the substitution of 4 of them led to variants with a decreased synthetic ability, 7 led to an increased ability and 3 gave no quantifiable yields.
As the rationalization of the results proved unsuccessful, it seems to be essentially impossible to identify by rational design the key positions to mutate. However, the methodology proposed in this work to create improved transglycosidases is much more rapid and efficient than directed evolution, which requires screening procedures on a large library of mutants. As it has been proved efficient for creating both α- (this work) and β-transglycosidases (Teze et al. 2014), it may be a general route to obtain transglycosidases rapidly.
Materials and methods
α-Gal-pNP and β-Gal-S-Ph were purchased from Carbosynth (Compton, UK), oligonucleotides from Eurofins (Luxembourg, Luxembourg) and enzymes from Fermentas (Waltham, Massachusetts). Other laboratory reagents were purchased from Sigma-Aldrich (St Louis, Missouri) unless otherwise indicated and used without further purification.
Site-directed mutagenesis
The QuikChange site-directed mutagenesis kit from Stratagene was used to make point mutations without any modifications: During polymerase chain reaction, Pfu polymerase extends the primers and generates a nicked plasmid with the desired mutation incorporated. Then the product is treated with Dpn I endonuclease, which digests the parental DNA. The nicked plasmid can then be transformed into XL-1 Blue competent cells. The 1.3-kb DNA fragments encoding the mutant AgaB enzymes were sequenced in both forward and reverse directions. Primers used for the site-directed mutagenesis are listed in Supplementary Data.
Protein expression and purification
Recombinant strains expressing AgaB genes were grown in 1 L of Luria-Bertani medium at 37°C overnight, centrifuged and resuspended in 35 mL of lysis buffer (0.1 M phosphate, pH 8, 0.5 M NaCl, 10 mM imidazole, 10 µg mL−1 DNAse I). After sonication and centrifugation, 6 × His-tagged proteins were purified by immobilized ion metal-affinity chromatography (IMAC): 250 µL of Ni-NTA Superflow (Qiagen) was added to the supernatant and stirred for 1 h at 4°C, then loaded onto a 10 mL column. The column was washed using 25 mL of washing buffer (0.1 M phosphate, pH 8, 0.5 M NaCl, 25 mM imidazole), then 5 mL of elution buffer was added (0.1 M phosphate, pH 8, 0.5 M NaCl, 250 mM imidazole). Purity of the final product was checked by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Enzyme concentrations were determined by UV absorbance at 280 nm (Abs 0.1% = 2.18 and 2.07 for the W mutants) using NanoDrop 1000 (Thermo Scientific)
Bioinformatic analysis
GH36 sequences were retrieved from the Uniprot database (Apweiler et al. 2004) and multiple sequence alignment (MSA) was performed with CLUSTALW (Chenna et al. 2003). To assess the degree of amino acid residue conservation, 205 different sequences >30% identical to AgaB after the MSA were considered. Hereafter, amino acid residues found conserved in >99.5% of these sequences are termed “highly conserved.”
Kinetic studies
All kinetic studies were performed with a TECAN Infinite M1000 spectrophotometer (TECAN France S.A.S.U., Lyon, France) at 30°C in phosphate buffer 0.1 M, pH 7. Initial reaction rates were calculated from the slope of the first-order plot of product concentration (pNP) against reaction time, then initial reaction rates were plotted against substrate concentration and the resulting curves were fitted to the hyperbolic equation v = Vmax ×[S] ×[E]/(KM + [S]) using Origin 7.0 (OriginLab) to obtain kcat and KM parameters. For most mutants hyperbolic curves were not observed since the kinetic of pNP release was the result of glycosylation, hydrolytic and transglycosylation reactions. The kcat/KM parameter was evaluated at low substrate concentrations, when kinetic can be approximate to the equation: v = kcat/KM ×[E] ×[S]. kcat/KM is then an apparent second-order rate constant (referred as the “catalytic efficiency”), which is determined by the initial slope of the curve V/[E] = f([S]). All measurements were made in triplicate with at least 12 different substrate concentrations ranging from 9.8 µM to 20 mM.
Transglysosylation studies
Screening of the transglycosylation efficiency of AgaB variants was achieved by following GalGal-pNP synthesis (Spangenberg et al. 2000). Synthetic yields with 10 mM α-Gal-pNP were determined by capillary electrophoresis (Beckman P/ACE System 5000 with an uncoated fused silica capillary, 47 cm) (Teze and Dion 2013; Teze et al. 2014). 200 µL of media containing 10 mM imidazole (used as an internal standard), 10 mM α-Gal-pNP and the enzyme (enzyme concentrations are indicated in Tables II–IV) was incubated at 30°C in phosphate buffer 0.1 M, pH 7, within the capillary electrophoresis apparatus and analyzed every 24 min for 2.8 h then each hour for the remaining 7 h. If specified, α-galactosyl-fluoride (α-Gal-F) 10 mM or β-galactosyl-azide (β-Gal-N3) 10 mM was used as a donor instead of α-Gal-pNP. If specified, 1-thio-β-d-galactopyranoside (Gal-S-Ph) 10 mM was used as an acceptor, in which case the given transglycosylation yields refer to the synthesis of phenyl-(α-d-galactopyranosyl)-(1,6)-1-thio-β-d-galactopyranoside (GalGal-S-Ph). Separations were performed at 17 kV with 50 mM Borax, pH 9.5, as running buffer. Donors (except α-Gal-F), acceptor and products were detected by UV absorbance at 214 nm and quantified by comparison with imidazole. Four experiments with the native enzyme and the N334A mutant gave maximum yields within a percent precision. As an example, all 15 electropherograms obtained for the best variant (N334T) study are presented in Supplementary Data. Due to its high thermostability, native enzyme and also AgaB variants are fully stable in the timescale of experiments.
Supplementary material
Funding
This work was supported by the Pays de la Loire and Bretagne regions (Glyconet Network) and the French Agence Nationale de la Recherche (ANR-10-BLAN-0718).
Conflict of interest statement
None declared.
Abbreviations
AgaB: Geobacillus stearothermophilus α-galactosidase B; GalGal-pNP: 4-nitrophenyl-(α-d-galactopyranosyl)-(1,6)-α-d-galactopyranoside; GalGal-S-Ph: phenyl-(α-d-galactopyranosyl)-(1,6)-1-thio-β-d-galactopyranoside; Gal-S-Ph: 1-thio-β-d-galactopyranoside; GH36: glycoside hydrolase family 36; TmαGal: Thermotoga maritima α-galactosidase; PDB, protein data bank; WT: wild type; α-Gal-F: α-Galactosyl-fluoride; α-Gal-pNP: 4-nitrophenyl-α-d-galactopyranosyl; β-Gal-N3: β-galactosyl-azide.
Acknowledgments
The authors are grateful to Genadi Chikov, Corentin Léger, Amina Fateh and Claude Solleux for their help and technical support.